By Gokhul Srinivasan, Sr. Partner Solutions Architect – AWS
By Cedric Druce, Director of Product Marketing – MemVerge
| MemVerge |
 |
Compute cost for next-generation sequencing (NGS) genomic pipelines can be high due to the data volume, algorithmic complexity, and need for high-performance computing resources. Cost optimization makes these pipelines accessible to researchers and can lead to faster and more efficient genomic data processing.
In this post, we explore how Sentieon improved its cost and performance using MemVerge’s Memory Machine Cloud (MMCloud), which streamlines the deployment of containerized applications on Amazon Elastic Compute Cloud (Amazon EC2) instances.
MemVerge is an AWS Specialization Partner and AWS Marketplace Seller with the Amazon EC2 service ready designation. MMCloud is a cloud automation platform designed for bioinformaticians and data scientists to easily run computational workflows on AWS.
Sentieon provides a commercial package of bioinformatics analysis tools for processing genomics data. The Sentieon Genomics tools are new implementations stressing computational efficiency, and accuracy of the algorithms underlying popular open-source tools such as Genomic Analytics ToolKit (GATK), Picard, and BWA-MEM. Sentieon Genomics tools show a 5-10x performance improvement over the equivalent open-source tools.
MMCloud includes a checkpoint/restore feature that automatically migrates running processes, from one Amazon EC2 instance to another, without losing the execution state. This feature dynamically right-sizes EC2 instances and protects against Amazon EC2 Spot instance reclaims.
The variant calling process identifies variants from genome sequence data. Variant calling pipelines need different CPU and memory capacity at different stages of the process. When Sentieon Genomics tools are used with MMCloud, each stage requires a different type of EC2 instance.
As the resource demands change, MMCloud migrates the running processes to an EC2 instance with the appropriate resources. This ensures the resources available to the job are not over- or under-provisioned—a capability not available from other cloud automation solutions, which provide horizontal scaling only.
This post compares the compute performance of a whole genome sequencing (WGS) pipeline, executed with and without MMCloud. In this example, the execution on MMCloud reduces wall clock time by 40% and cost by 34%, respectively. The results are generalizable as any pipeline (like a machine learning model) where the peak resource demands (CPU and/or memory) are greater than the average demands should realize the same benefits from using MMCloud.
WGS Pipeline
Sentieon publishes a catalog of example pipelines, including a germline variant calling pipeline (using Sentieon’s DNAscope product) for whole-genome samples. This WGS pipeline was used in the tests described here and comprises four steps as shown in table below.
| Step | Function | Description | Output | Example of Open-Source Analog |
| 1 | Data input | Dataset (FASTA files) loaded into local storage | Dataset available on local disk | N/A |
| 2 | Sequencing mapping | Align local sequence to reference genome | Coordinate-sorted BAM file | BWA-MEM |
| 3 | De-duplication | Remove duplicate reads | De-duplicated BAM file | Samtools or Picard (included in GATK) |
| 4 | Variant calling | Haplotype-aware variant detection | Variant Call Format (VCF) file | GATK |
Performance Analysis
In terms of resource requirements, the four steps of the WGS pipeline fall into the following three periods (numbered steps refer above table in WGS pipeline):
- Period 1: Data input (Step 1)
- Period 2: Sequence mapping (Step 2)
- Period 3: GATK procedures (Steps 3 and 4)
The Sentieon WGS pipeline runs as a single-server application; that is, a shell script configures multiple compute steps on a single EC2 instance. For the performance tests, the WGS pipeline runs on one (for the baseline) or more than one (when used with MMCloud) On-Demand EC2 instances.
The latest build of the human reference genome (known as Hg38) is used as the reference FASTA file for all tests. Data is public and available from multiple sites; for example, as a GitHub source.
Baseline
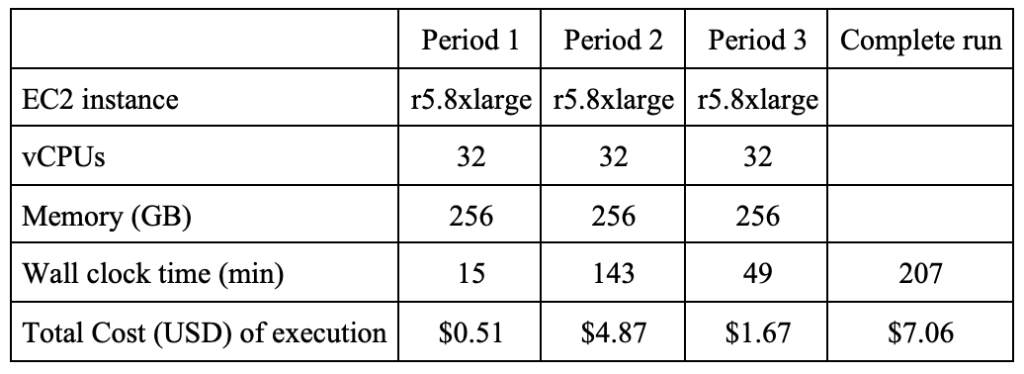
To establish the baseline, the WGS pipeline was run on a single r5.8xlarge instance (32 vCPUs and 256 GB memory). Even though the r5.4xlarge instance (16 vCPUs and 128 GB memory) meets the resource requirements, the r5.8xlarge instance was available at a lower price than the r5.4xlarge instance.
MMCloud includes both a command line interface (CLI) and web interface to submit and manage jobs. The web interface provides a graphical display showing how the resource demands (CPU, memory, and network) change as a job executes. The profile for the baseline is shown below.
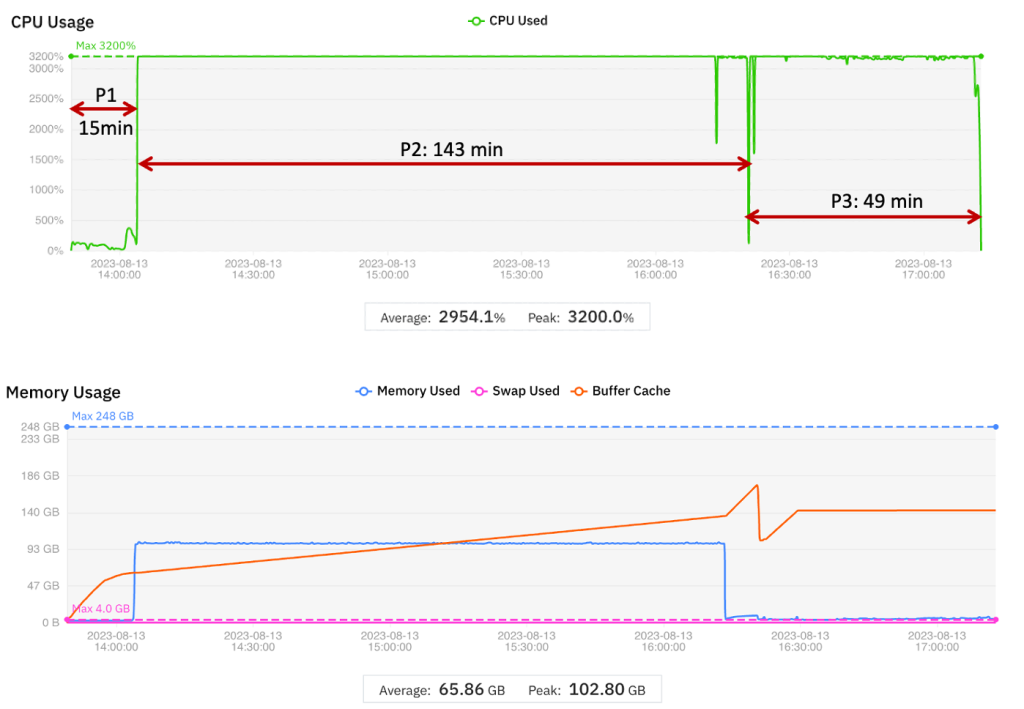
Figure 1 – CPU and memory utilization for the baseline.
The display of CPU utilization (summed over 32 vCPUs, so the theoretical maximum is 3,200%) is annotated to show the wall clock times corresponding to Periods 1 through 3, respectively. Numerical results are summarized in the following table.
Figure 2 – Wall clock time and cost for the baseline.
Discussion
The baseline run shows three distinct behaviors.
- In Period 1, vCPUs are underutilized – total CPU load is 100% or less (maximum possible is 3,200%). Memory utilization is also low (4 GB) compared to the 256 GB available.
- In Period 2, the multi-threaded implementation uses all 32 vCPUs. Memory utilization is high, although the peak (103 GB) is less than the 256 GB capacity.
- In Period 3, all 32 vCPUs are used although the memory utilization is low (comparable to Period 1).
The CPU and memory utilization profiles indicate areas where performance and cost can be optimized. The ratio of peak memory usage to average memory usage is large, which means that, for part of the run, memory is over-provisioned even when CPU utilization is high. If the EC2 instance is sized to the average memory used, then the job would either run slowly (because of excessive memory swapping) or fail due to out-of-memory (OOM) errors.
The CPU utilization in Period 1 shows the pipeline does not employ more than 4 vCPUs at any time during this period, which indicates that an EC2 instance with 4 vCPUs and 32 GB memory can be used without incurring a performance penalty. In contrast, the CPU utilization in Period 2 indicates the Sentieon software uses all available vCPUs for the duration of Period 2, and performance can be improved by using an EC2 instance with more than 32 vCPUs.
WGS Pipeline with MMCloud
With MMCloud, job migration (moving of running processes to a new EC2 instance) can be initiated manually (using the CLI or web interface), programmatically (inserting CLI commands into the job script), or by policy (crossing CPU and/or memory utilization thresholds).
To investigate the effect of MMCloud on WGS pipeline performance, migration events were triggered programmatically by inserting MMCloud CLI commands into the WGS pipeline shell script at the junctions where one period ends and a new period starts.
A key factor to optimize software performance is ensuring that processes are not delayed because they are waiting for resources to free up. Sentieon provides a CLI to increase or decrease the maximum number of process threads (the number of process threads are not visible to MMCloud).
In this test, the maximum number of threads is increased to 64 to match the number of vCPUs available in Period 2. If the maximum number of threads is more than the number of vCPUs, the effect on performance is neutral or negative (because of excessive context switching). For this reason, the baseline test used 32 as the maximum number of threads.
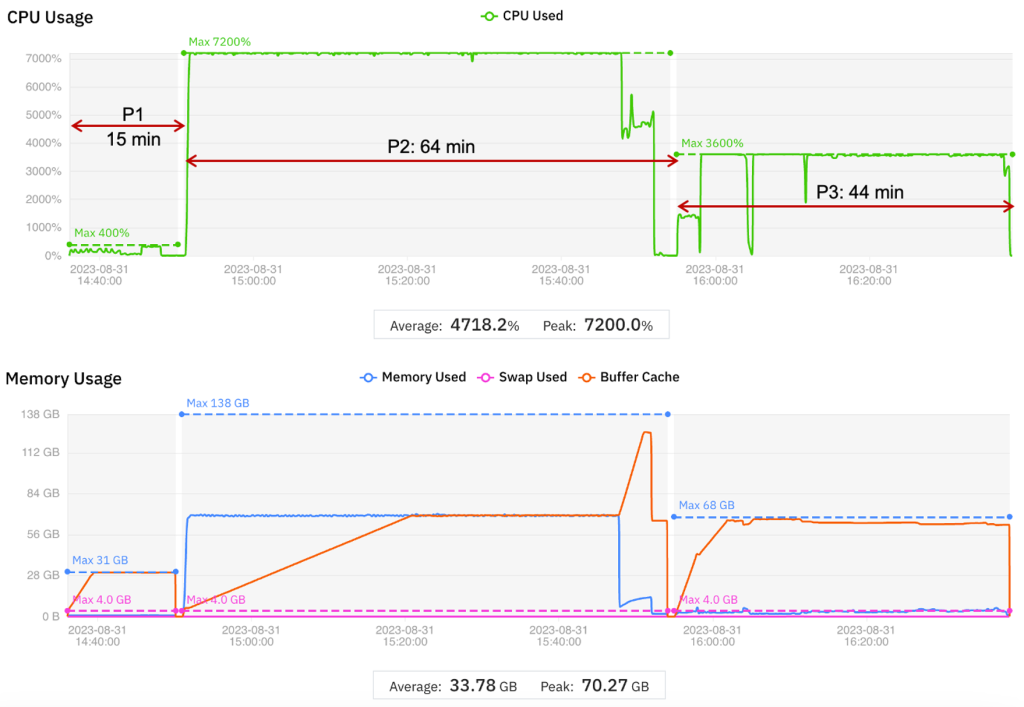
To optimize cost and performance, the following procedure was followed:
- Increase the maximum number of threads to 64.
- Start Period 1 with a small EC2 instance (4 vCPUs and 32 GB memory) to decrease cost.
- At the start of Period 2, migrate to a large EC2 instance (64 vCPUs and 256 GB memory) to decrease wall clock time.
- At the start of Period 3, decrease the maximum number of threads to 32 and migrate to an EC2 instance with 32 vCPUs and 64 GB memory (to decrease cost).
The results are displayed graphically in the figure below. The time taken to migrate to a new virtual machine is included in the wall clock time.
Figure 3 – Graphical display of job migration to optimize cost and performance.
The comparison with the baseline is shown in the below table.
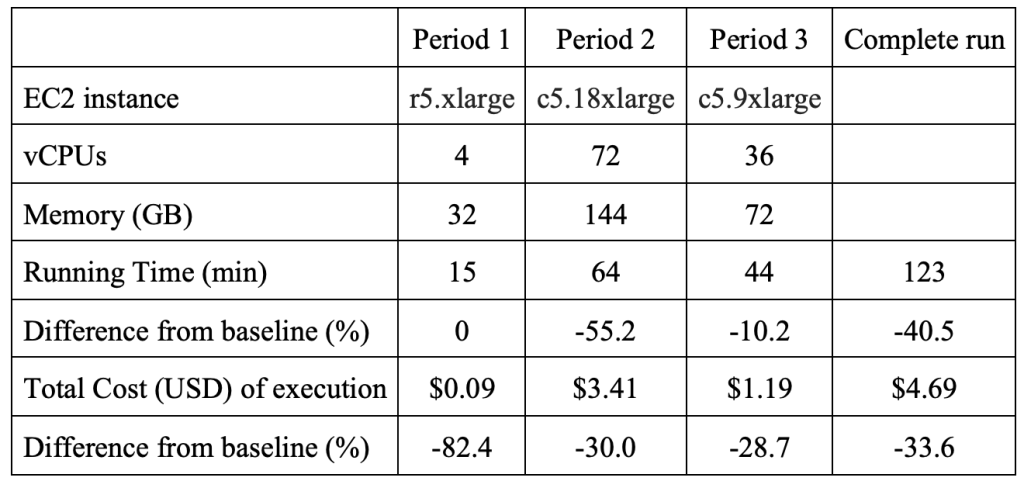
Figure 4 – Comparison of cost and performance optimization test with baseline.
Discussion
There are two options for specifying the EC2 instance when submitting a job to MMCloud:
- Specify the instance type by name; for example, r5.8xlarge.
- Specify a range of vCPUs and range of memory capacity; for example, the number of vCPUs (or memory) must be at least 32 (or 128 GB).
With the second option, MMCloud selects the EC2 instance with the lowest price that meets the vCPU and memory requirements.
In this test, MMCloud searched for an EC2 instance with at least 64 vCPUs and 128 GB memory and found an instance with 72 vCPUs and 144 GB memory. With this EC2 instance, Period 2 completed in less than half the wall clock time while reducing Period 2 cost by 30% (compared to the baseline).
Test Summary
Tests comparing the performance of a Sentieon WGS pipeline, with and without MMCloud, show that with MMCloud both wall clock time and cost decrease.
Compared to the baseline on a single (unchanging) EC2 instance, the MMCloud inclusion shows a 40% decrease in wall clock time and 34% reduction in cost. Cost reduction reflects the prices of Amazon EC2 instances (On-Demand and Spot) at the time the tests ran. Prices are subject to market fluctuations and results vary if tests are repeated.
Customers can realize additional cost savings by using Spot instances instead of On-Demand instances and using MMCloud’s checkpoint/restore feature to handle Spot instance reclaim events without losing state. This report did not investigate this aspect.
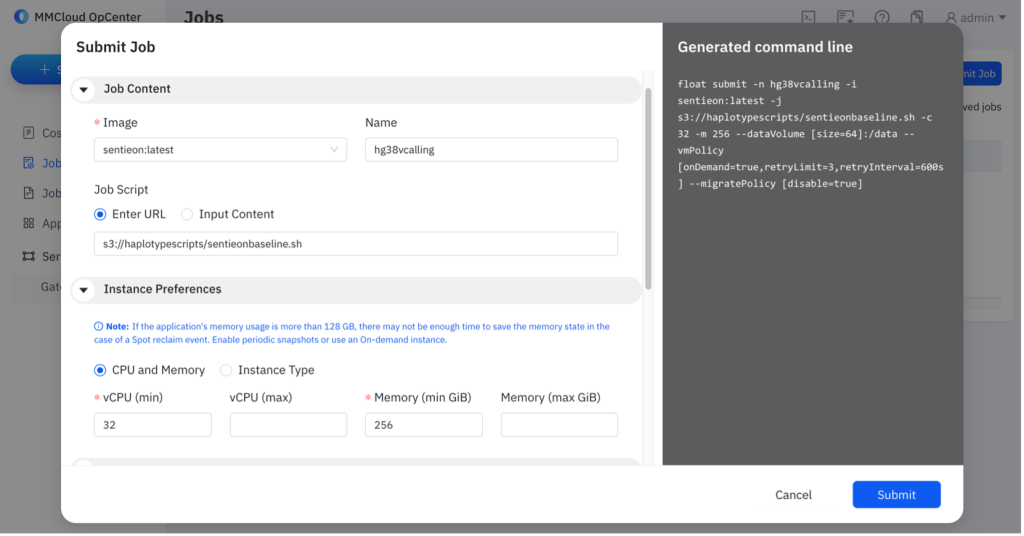
MMCloud is software used to streamline the deployment and management of containerized applications (both batch and interactive) in EC2 instances. At the core of MMCloud is the Operations Center (OpCenter), which is a containerized application deployed by the user inside the virtual private cloud (VPC) of their AWS account.
The user submits jobs to the OpCenter using the MMCloud CLI or web interface. The OpCenter takes care of the rest: identifying and instantiating AWS resources (and deleting resources when they’re no longer needed). Each job runs in its own container in its own EC2 instance to maximize security and data privacy. For users who are new to AWS, MMCloud provides a simple way to deploy containerized applications at scale.
Bioinformatics or machine learning computational pipelines have complex analyses comprising a series of tasks, each of which may involve different software and dependencies. From the MMCloud perspective, each task in pipeline is a separate batch job, deployed and managed by the OpCenter.
In Sentieon’s case, the sequence of tasks in the pipeline is defined by a shell script. In other cases, a workflow manager, such as Nextflow or Cromwell, is used to drive the pipeline.
Figure 5 – Submitting a job using the MMCloud web interface.
Conclusion
Sentieon genomics tools decrease wall clock times for executing a wide range of bioinformatic pipelines when compared to open-source tools. MemVerge’s MMCloud offering is available on AWS Marketplace for streamlining the deployment of computational pipelines on EC2 instances.
Tests show reduction in both wall clock time and cost when using Sentieon genomics tools in combination with MMCloud to dynamically right-size EC2 instances.
The results apply to any data computational pipeline where the resource demands vary widely, and for extended periods of time. MMCloud ensures that the EC2 instance on which the application is running is not over- or under-provisioned.
Multi-threaded software implementations, like Sentieon genomics tools, take advantage of the resources that are available. When used together, the outcome is performance improvement and cost reduction.
MemVerge – AWS Partner Spotlight
MemVerge is an AWS Specialization Partner whose cloud automation platform (MMCloud) is designed for bioinformaticians and data scientists to easily run computational workflows on AWS.
Contact MemVerge | Partner Overview | AWS Marketplace